6.2: Mass Spectrometer Ionization Techniques for Membrane Proteins
- Page ID
- 17671
Mass spectrometry is a technique in which charged particles from a sample are analyzed in order to obtain structural and compositional information about the sample. Mass spectrometry is particularly relevant when studying proteins. With this instrument, the determination of protein structure, function, and interactions is possible. A mass spectrometer can also detect post translational modifications on complex mixtures, perform both quantitative and qualitative analysis, and monitor enzyme reactions, chemical modification, and digestion pathways. 1 To date, two main ionization methods are used for protein analysis, electron Ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). These two ionization methods will be detailed in this page along with methods used prior to these discoveries such as electron bombardment (EI) and fast atom bombardment (FAB). A few applications of Mass spectrometry for membrane protein analysis will also be covered.
Introduction to Mass Spectrometry
A diagram of a conventional mass spectrometer is depicted in Figure \(\PageIndex{1}\).
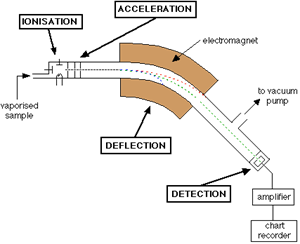
As shown, the mass spectrometer consists of a sample input, an ionization chamber, accelerator, defector, detector, and an amplifier. In brief, the sample of interest is injected into the mass spec and ionized to form charged particles. The charged particles are then accelerated by increasing the kinetic energy of the particles. As shown in the Figure, there are three plates with slits in the acceleration region; these plates all vary in potential allowing for the particles to increase in kinetic energy and form a finely tuned beam as they pass through each slit. This beam consists of a mixture of different ions before hitting the deflector. The deflector takes this mixture and separates the ions based on mass to charge ratio (m/z); this is done by applying a magnetic field. Figure \(\PageIndex{2}\) presents a diagram of what occurs in the deflector.

Different ions interact differently with the magnetic field. In Figure \(\PageIndex{2}\) for example, the ions from steam B are ions that have a low m/z ratio. This means that the ions that compose this beam either have a low mass or a high charge or both. On the other hand ions from stream C hardly interact with the magnetic field applied; meaning they have a high m/z ratio. The ions in steam B, however, interact with the magnetic field in such a way that the ions pass through to the detector. The magnetic field can be altered to select a desired m/z range. For example, if one were to want to detect the higher mass ranges (steam C), the magnetic field would need to be larger in order to deflect the ions more. Equations 1 (Lorentz Force) and 2 (Newton’s second law of motion) are crucial in understanding how partials move in an electric or magnetic field.
\[F=Q(E+v \times B) \label{1}\]
\[F=m a \label{2}\]
Where \(Q\) is the ion charge, \(F\) is the force applied on the ion, \(v \times B\) is the cross product of the velocity and magnetic field, \(m\) is the mass and a is the acceleration.
Equating the two equations then gives the following differential (Equation \ref{3})
\[(m / Q) a=E+v x B \label{3}\]
Once the selected ions pass the deflector the ions travel towards a detector were there m/z is recorded. In short, when the ions strike the metal detector, a current from the movement of electrons is produced which can then be recorded as a signal. Because the electrons affected by this collision are typically very few, amplification of the signal is almost always necessary. Ions in the gas phase are typically very reactive and have a short lifetime thus it is important that the instrument in run in high vacuum (typically from 10−3 torr to 10−6 torr pressure).
There are several different mass spectrometers commercially available that tailor to different needs, such as quantifying data, qualifying data, protein sample analysis, small sample analysis, etc. Ionization techniques have been key to determining what types of samples can be analyzed by mass spectrometry.3
Hard and Soft Ionization
Before going into detail about the different ionizations methods it is important to understand two main categories that ionization falls under, hard and soft ionization.4
- Hard ionization- hard ionization evokes larger amounts of energy to the sample of interest in order to ionize the sample. Due to the larger amount of energy the bonds within the molecule tend to break more, resulting in an increase in fragmentation. Hard ionization techniques typically yield in a larger number of lower mass fragments as oppose to higher mass.
- Soft ionization- Soft ionization methods use smaller amounts of energy to ionize the sample, causing a decrease in fragmentation. This technique yields a larger amount of high mass fragments.
Electron Ionization (EI)
Theory
Electron Ionization (EI) was one of the first ionization techniques developed for mass spectrometry. 5 EI is a hard ionization method where a beam high-energy electron is used to bombard a sample of interest to produce multiple ions. As previously mentioned the use of high-energy particles leads to extensive fragmentation, which can be useful for structural characterization. A diagram of EI is shown is Figure \(\PageIndex{3}\).

As shown, the apparatus used to ionize molecules via EI consist of a source block, sample inlet, repeller, magnets, filament, electron trap, and an exit plat at ground potential. The source block is made out of metal heated to 3000C to prevent the sample from condensing. The sample must first be introduced into the source block in the gas phase. This can be accomplished by either “boiling off” the sample from a probe via thermal desorption or by introducing of a gas through a capillary 7 . Once these neutral gas phase particles enter the source block, they are bombarded by a beam of electrons where the electrons excite the molecule and cause the molecule to eject an electron from the highest occupied molecular orbital (HOMO). This process results in ejection ionization and molecular fragmentation. The electron ionization process that occurs is described below: 5
\[M+e^{-} \rightarrow M^{+\cdot}+2 e^{-}\]
where \(M\) is the analyte molecule, \(e-\) is the electron, and \(M^{+\cdot}\) is the resulting positively charged molecular ion.
The electron source (cathode) is typically a thin filament of tungsten or rhenium wire that is heated up to an incandescent temperature to produce electrons termed thermionic emission. These electrons must have enough energy to overcome the ionization energy of the sample molecules. A potential of 70 V is applied between the cathode and the source block in order to accelerate the electrons to 70 eV. At approximately 70 eV, the de Broglie wavelength of the electrons equals the length of a typical organic molecular bond. The energy transfer to organic analyte molecules is maximized thus leading to the strongest possible ionization and fragmentation.7 A slightly positive electron trap plate (anode) is placed at the opposite end of the source block. The excess electrons that did not participate in the ionization of the sample are collected here. A small magnetic field is applied parallel to the path of the electrons, by doing so the electrons travel in a helical path and thus increases their path length which in turn increases the probability of interaction between the sample and the electron. The resulting positive charged sample molecules are accelerated by the repeller into the accelerating region. By maintaining the exit slit at ground potential, the ions enter the mass analyzer at a fixed kinetic energy. 7
The fragmentation and ionization process can be described using the Born Oppenheimer potential curves depicted in Figure \(\PageIndex{4}\).

The Figure shows the Born Oppenheimer potential curves of a methanol molecule before and after electron impact. The red arrow depicts the energy transition between the neutral molecule and the ionized molecule without sample fragmentation. However with this method this is typically not the case. Due to the high energy of the incident electron, dissociation of the molecule is more likely. The energy transition described is illustrated by the blue arrow.
Because this is a hard ionization technique, EI is good for polypeptide sequencing but it is very difficult to obtain information of large proteins and their interaction pathways. As we will later see, along with all ionization techniques come advantages and disadvantages.
a) Advantages
- High fragmentation can aid in molecular characterization
- Sample can be solid, liquid, or gas
- High fragmentation allows for a well defined fingerprint spectra
- Can be equipped to many different mass analyzers including time-of-flight, GC-MS, LC-MS etc.
- Fast and easy (typically first method of choice when screening samples) 5
b) Disadvantages
- Characterization of large molecules is nearly impossible
- Sample must be volatile
- Multiple charges can result in very confusing mass spectra.
- Low mass ranges typically less than 600 Da. ( Proteins can be in the kDa range)
- Extensive fragmentation can make it difficult to interpret data.
- Only positive ions are formed
Application to Proteins
Electron Ionization Mass Spectrometry (EI-MS) has be used in protein turn-over research. The rate of protein synthesis in human subjects is of great interest especially in this era were being "healthy" and "fit" and gaining lean muscle is of much interest. A study conducted in 1992 by Calder AG reports using EI-MS to determine the enrichment of amino acids incorporated into tissue protein during studies of human protein synthesis.20 This was done by measuring the d-5 enrichment from the conversion of phenylalanine to phenylethylamine via EI-MS. The method of analysis proved to be very efficient for this task because detection of both free and protein-bound d-phenylalanine were detected simultaneously and only 1 mg of protein was needed to conduct the study.20 However, EI-MS is not commonly used anymore for protein analysis because, since this is a hard ionization method, low mass ranges typically less than 600 Da are detected and Proteins are typically in the kDa range.
Fast Atom Bombardment (FAB)
Theory
Prior to the 1980s, EI was the main source of ionization for mass analysis. As previously discussed, one major limitation to all chemist and biochemists using this technique was the inability to ionize larger bioorganic molecules such as polypeptides. This limitation motivated scientist to develop new and improved ionization techniques that would allow for these larger molecules to be ionized and detected. Fast atom bombardment (FAB), MALDI, and ESI were all designed to circumvent this limitation. These techniques revolutionized biomolecular analyses.7Fast atom bombardment (FAB) is a soft ionization technique in which a sequence of high-energy atoms that strike a surface to produce ions. Figure \(\PageIndex{5}\) present a diagram of how FAB functions.

As shown a stream of fast particles, typically an inert gas such as Argon or Xenon, bombards a sample of interest emitting analyte of various masses from the plum.
In its natural state, Argon and Xenon are relatively slow moving particles. In order to accelerate these molecules to be used as a beam source, the gas must first be ionized itself, this is done by colliding small particles or atoms. Once ionized, the gasses can then be accelerated to a certain potential. When in motion, these fast moving ions become neutralized by the dense cloud of excess natural gas surrounding them in the collision cell, this process in known as charge exchange. This process can be expressed below: 9
\[\begin{align} Ar &\rightarrow Ar^{+} + e^{-} \\[4pt] Ar^{+} &\rightarrow Ar^{+}(\text {fast }) \\[4pt] A r^{+}(fast) + Ar &\rightarrow Ar(fast) + Ar^{+} \end{align}\]
Because FAB is a soft ionization technique, most of these ions being ejected by the bombardment of these neutral inert atoms will have high masses. The sample itself is not just consistent of the analyte, like Matrix Assisted Laser Distortion Ionization that will be discussed later in this article; FAB uses a matrix to aid in ionization. FAB operates using a liquid matrix, where the sample and a large excess of matrix are mixed. The sample-matrix mixture is placed on a sample plate typically using an insertion probe. Having a liquid matrix services FAB by reducing damage made to the sample by the impact of the ion beam, constantly replenishing the surface with new sample after being bombarded with incident beam, and keeping the sample from aggregating. 10Below is a list of factors to consider when selecting a matrix for a sample:
- The sample must be soluble in the matrix
- Matrix cannot react with sample ions
- Matrix must have low volatility under vacuum in order to maintain a liquid
One of the most commonly used liquid matrices for FAB ionization is glycerol and m-nitrobenzyl alcohol (NBA). 11 Once the sample is prepared and placed on the probe, the instrument is placed under high vacuum (typically from 10−3 torr to 10−6 torr pressure) to insure that no other molecules will contaminate the sample or react with the very reactive ions being produce. The sample is then irradiated with a continuous beam of high energy atoms (from 4000 to 10,000 electron volts) producing ions that can be analyzed using a mass spectrometer.10 Listed below are advantages and disadvantages with using FAB as an ionization method.
a) Advantages
- · Because it is a soft ionization technique samples with large masses can be analyzed
- · Able to produce large biological samples such as small proteins, peptides, DNA, etc.
- · Can analyze polar, ionic, and thermally and energetically liable samples
- · Can be equipped to a quadruple
- · Allows for less complicated spectra because of its tendency to produce mostly singly charged particles.
b) Disadvantages
· Characterization using smaller fragments can be very difficult
- · Usually form single charged particles decreasing the effective range of the mass analyzers.
- · Low sensitivity compared to MALDI
- · Lower mass ranges compared to other Ionization techniques seen later in this report (~300-6000 Da)
- · Sample must be soluble in the matrix
- · Salt contamination can suppress molecular ion formation
- · Typically spectra have high chemical background
Applications to Proteins
Theoretically FAB would be very useful for protein membranes because, as previously mentioned, the matrix/sample mixture is typically glycerol and in this matrix the proteins would rise to the surface of the mixture. 21 Compounds at the surface of the matrix are readily ionizable.21 However, many studies show an decrease in ion current with increase mass of sample, meaning large proteins (greater than 3000 Da) would be difficult to ionize. 21 Previously FAB has been used for the annalyis of large peptides and smaller proteins such as Proinsulin23, glucagon24, melittin25, adrenocorticotropic hormone26 and has also been used to sequence small peptides. 27
Electrospray Ionization (ESI)
Theory
The need for a new and improved ionization method became apparent when scientist started studying proteins in greater depths. Before electrospray ionization, ionization methods such as fast atom bombardment were used to ionize small proteins. However, as previously discussed one great limitation in this ionization method is the inability to ionize large molecules such as large proteins at high yields. These limitations were circumvented when Fenn developed a technique known as Electrospray Ionization in 1989.12 ESI is a soft ionization technique in which ions are ejected from charged vapor droplets to produce an aerosol. Figure \(\PageIndex{6}\) presents a diagram of an ESI probe.
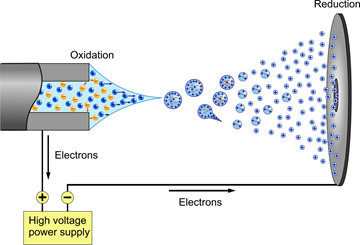
A dilute sample is introduced by a mechanical syringe pump through a hypodermic needle at low flow rate of typically 1–20 μL/min.12 The capillary is typically heated to 100–300°C in order for the ions to be completely desolvated. As shown in the diagram above, a voltage (~4000 volts) is applied between the capillary containing the sample (left) and the sample plate (right). The voltage causes charge separation at the tip of the capillary. This charge separating and the full of the electric field on the positive ions overcomes the surface tension of the liquid thus forming a Taylor cone at the end of the capillary that contains an excess of charge. 14 The tipoff the cone being the least stable point, elongates into a liquid filament. For the case above (Positive Ion Electrospray), the positively charged ions are arranging at the tip of the Taylor cone. This cone is formed to relieve columbic repulsion at the capillary tip once Rayleigh limit of surface tension is reached. From this cone a stream of droplets form. As the droplets travel to the sample plate the solvent in the droplet starts to evaporate. This process can be aided by using an inert gas such as N2 to help nebulize the droplets. As the solvent evaporates the charge density becomes more concentrated and in turn causes like charges to repel each other (Coulombic effect). This effect can be described using Equation 4 (Coulombs law).

The repulsion of these charges causes and increases in surface tension of the droplet; when the droplet can no longer hold, the Rayleigh limit is exceeded.15 This process reoccurs as more solvent evaporates within each droplet until the individual ions reach the sample plate; by this point all ions should be separated, this process in known as the Dole charge residue model. Equation 5 describes the maximum charge a droplet can carry before the Rayleigh limit of surface tension is reached.16

However, there is another theory as to what happens when a droplet is traveling to the sample plate, known as the Iribarne Thompson ionization theory. Here, it is said that a point is reached in which the repulsion between the ions is so great that it is thermodynamically unstable for the ions to stay together and thus it is favorable for hydrated ions to be liberated from the droplet into the gas phase. When the droplet travels to the sample plate the radius of the droplet becomes smaller and smaller due to the evaporation of the solvent. Iribarne Thompson ionization theory states that there is a point where the radius is so small (less than 10nm) that the ion emission dominates over Rayleigh fission. 14 This process does not require that a very small droplet contains only a single ion as seen in the Charge Residue model.12 Iribarne Thompson ionization theory states that the using Equation 6.12

.Common solvents used in ESI include a mixture of water with a volatile organic compound such as methanol. In order to decrease the initial droplet size small amounts of acetic acid is commonly added. It is important to have the correct solvent in order to optimize the ionization yield of the gas molecules. As previously mentioned, a Taylor cone is formed at the end of the capillary tip that is formed from overcoming of the surface tension of the liquid. The electric field required to onset the Taylor cone providing a steam of droplets is described using Equation 7. 14
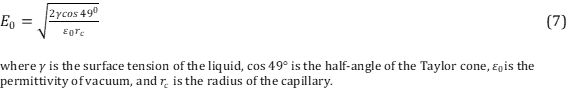
Notice the dependency of the surface tension of the liquid on the electric field. Liquids with high surface tensions will require higher electric fields.
The ionization of the ions in the charged droplet is dependent on the presence of charges that comes from the separation of positive and negative electrolyte ions in the Taylor cone. Thus, it can be said that concentrations of these electrolytes play a great role in optimizing the ionization of molecules. It has been shows that a minimum concentration of 10-5 M concentration is required.
Along with all ionization techniques, ESI comes with advantages and disadvantages to using this technique.
c) Advantages
- · Because it is a soft ionization technique samples with large masses can be analyzed
- · Able to analyze large biological samples such as proteins, peptides, DNA, etc.
- · Can be equipped to a quadruple, ion trap, and liquid chromatography
- · No matrix interference or limitation
- · Multiple charging allows for analysis of high mass ions with a relatively low m/z range instrument
d) Disadvantages
- · Characterization using smaller fragments can be very difficult
- · Difficulty with analyzing mixtures
- · Multiple charges can result in very confusing mass spectra.
- · Low-high mass ranges typically less than 200,000 Da.
- · Does not work well for non volatile salts (buffers)
- · Not useful for non-polar compounds
Applications to Proteins
ESI is a very common and useful tool for protein analysis mainly because it is able to ionize proteins without denaturing them, meaning non-covalent, receptor ligand complexes remain intact. This is because ESI allows for multiple charge states, meaning large proteins can be ionized and detected allowing ESI to be used for a wide range of applications.Characterization of Hemoglobin variants in blood have always been of interest in the clinical setting; however in the past it was very time consuming and challenging with other methods . With ESI-MS it is now possible to analyze over 250 samples in no more than 2 days. 28 ESI is also commonly used for amino acid profiling in order to detect major diseases such as phenylketonuria , maple syrup urine disease, homocystinuria, and galactosaemia. 28
Matrix Assisted Laser Distortion Ionization (MALDI)
Theory
Matrix-assisted Laser Desorption/Ionization (MALDI) along with electrospray ionization, are now among the leading ionization methods for nonvolatile, high molecular weight compounds. MALDI specifically has been widely used to analyze, nonvolatile biomolecules, in particular peptides, proteins, oligonucleotides, and oligosaccharides. 17
MALDI is a soft ionization technique in which a laser energy-absorbing matrix is used to create ions. Figure \(\PageIndex{7}\) presents a diagram of how MALDI functions.

A crucial step in utilized MALDI technique is the formation of the matrix-analyte cocrystal. The composition of the Matrix is a key difference between FAB ionization and MALDI. As previously mentioned FAB operates by utilizing a liquid matrix while MALDI uses a cocrystalline matrix. The cocrysal is made by dissolving analyte into a solution of the matrix compound. The solution is then spotted on the target plate, and once the solvent evaporates a mixture of analyte and the matrix compound is left. As shown, the target plate holds the sample (orange) and matrix compound (green). The matrix is in very large excess to insure that it gets desorbed with the target sample of interest.
One of the key components that drive the ionization of molecules in MALDI is the matrix the sample is placed in. The matrix compounds used in MALDI are generally low in mass; this is to ensure that once the laser strikes the sample the matrix is easy vaporized into the gas phase. In order for the matrix to have high enough energy to transfer energy to the analyte, it is important that the compound is able to have high UV absorbance. Typically highly polar analytes work best with highly polar matrices, and similarly nonpolar analytes work better combined with nonpolar matrices.18 Below is a table of commonly used matrices for different analyte.
|
Compound |
Acronym |
Application to |
|---|---|---|
|
Nicotinic acid |
NA |
Peptides, proteins |
|
Picolinic acid |
PA |
Oligonucleotides, DNA |
|
3-Hydroxypicolinic acid |
HPA, 3-HPA |
Oligonucleotides, DNA |
|
3-Aminopicolinic acid |
3-APA |
Oligonucleotides, DNA |
|
6-Aza-2-thiothymine |
ATT |
Oligonucleotides, DNA |
|
2,5-Dihydroxybenzoic acid |
DHB |
Proteins, oligosaccharides |
|
DHB-based mixtures |
DHB/XY and super-DHB |
Proteins, oligosaccharides |
|
3-Aminoquinoline |
3-AQ |
Oligosaccharides |
|
α-Cyano-4-hydroxycinnamic acid |
α-CHC, α-CHCA, 4-HCCA, CHCA |
Peptides, smaller proteins, triacylglycerols, numerous other compounds |
|
4-Chloro-α-cyano-cinnamic acid |
ClCCA |
Peptides |
|
3,5-Dimethoxy-4-hydroxycinnamic acid |
SA |
Proteins |
|
2-(4-Hydroxyphenylazo) benzoic acid |
HABA |
Peptides, proteins, glycoproteins, polystyrene |
|
2-Mercaptobenzothiazole |
MBT |
Peptides, proteins, synthetic polymers |
|
5-Chloro-2-mercaptobenzothiazole |
CMBT |
Glycopeptides, phosphopeptides, and proteins |
|
2,6-Dihydroxyacetophenone |
DHAP |
Glycopeptides, phosphopeptides, proteins |
|
2,4,6-Trihydroxyacetophenone |
THAP |
Solid-supported oligonucleotides |
|
Dithranol (1,8,9-anthracenetriol) |
None |
Synthetic polymers |
|
9-Nitroanthracene |
9-NA |
Fullerenes and derivatives |
|
Benzo[a]pyrene |
None |
Fullerenes and derivatives |
|
2-[(2E)-3-(4-tert-Butylphenyl)-2-methylprop-2-enylidene]malonitrile |
DCTB |
Oligomers, polymers, dendrimers, small molecules |
Once the sample is made, a high-energy laser pulse (typically UV) is applied to the sample creating a plume of ions that can then be analyzed. The type of laser used is another key differences between FAB and MADI, while FAB uses a series of atoms to bombard the sample; MADI operates using a beam of ions. Some common lasers used include a nitrogen laser (λ=337 nm) and a frequency tripled Nd-YAG laser (λ=355 nm). 11After the laser marks the sample, the irradiated spot is heated and becomes vibrationally excited. Desorption of excited molecules occurs where the molecules (consisting of the analyte and matrix) leave the target plate in the gas phase. The exact mechanism of this ablation is not know, however it is theorized that this occurs either because of sublimation after the excitation or pressure caused by rapid expansion of the crystal lattice. 11 During this process, the matrix molecules collide with the molecule of interest and protonate or deprotonate the molecule creating an ion. The ion is then able to travel to the mass analyzer. Although the exact mechanism of this ablation is not known there have been proposed mechanisms as to what could potentially be occurring when the sample is photionized. Below is a schematic of photoionized reaction pathways proposed by Ehring, Karas, and Hillenkamp in 1992 (Figure \(\PageIndex{8}\)).

Along with all ionization techniques, MSI comes with advantages and disadvantages to using this technique. 19
a) Advantages
- · Because it is a soft ionization technique samples with large masses can be analyzed
- · Able to analyze large biological samples such as proteins, peptides, DNA, etc.
- · Can be equipped to a quadruple which in turn leads to high accuracy when determining mass of sample since the quadruple m/z range is selective and relatively small.
- · Allows for less complicated spectra because of its tendency to produce mostly singly charged particles.
- · Can be used to image samples
- · Very high masses range typically less than 500,000 Da.
- · Tolerance of salts
- · Can be used to analyze complex mixtures
b) Disadvantages
- · Characterization using smaller fragments can be very difficult
- · Usually form single charged particles decreasing the effective range of the mass analyzers.
- · Contamination of sample can have a significant effect of making the crystal matrix.
- · Matrix could cause high background noise
- · Possibility of photodegradation by laser desorption/ionization
Applications to Proteins
MALDI, along side ESI, is also a very commonly used ionization method for protein analysis. Because MALDI is a soft ionization method, the ions produced have low energy allowing the production and detection of intact/low fragmented molecules. The production of intact molecules allows for protein identification and characterization. Once the protein is denatured, MALDI can also be used for protein sequencing by peptide mass fingerprinting.
One major advantage to MALDI is the ability to image proteins. MALDI Imaging can provide local molecular composition, relative abundance and spatial distribution of peptides and proteins in membranes.
Concluding Remarks
Ionization is a very important component of mass spectrometry and, as shown, is the key to determining what types of samples can be analyzed. As discussed there are important parameters and limitations associated with every ionization method, and understanding the limitations associated with each one is necessary in order to select the correct method when trying to analyze a sample. To date the ionization yields of large molecular samples such as proteins is relatively low (1/1000 molecules) limiting the sensitivity of mass spectrometers. 14 Novel ionization techniques are being sought out for that will improve upon this great limitation. Table 2 found summarizes and compares the ionization techniques visited in this article. 24
Table 2. Summary of ionization techniques visited in this paper.
|
Ionization source |
Typical mass range (Da) |
Matrix interference |
Degradation |
Complex mixtures |
LC/MS amenable |
Sensitivity |
|
Electron ionization (EI) |
500 |
NO |
NONE |
Limited unless used with GC/MS |
Very Limited |
Picomole |
|
Comments: Good sensitivity; unique fragmentation data generated; National Institute of Science and Technology (NIST) database (>100,000 compounds) available to compare fragmentation data; thermal decomposition a major problem for biomolecules; Limited mass range due to thermal desorption requirement. |
||||||
|
FAB |
7,000 |
YES |
Matrix reactions and some thermal degradation |
Somewhat amenable |
Very Limited |
Nanomole |
|
Comments: Relatively insensitive; little fragmentation; soft ionization; high salt tolerance to 0.01 M, solubility with matrix required |
||||||
|
Electrospray Ionization (ESI) |
70,000 |
NO |
Thermal |
Limited |
Excellent |
High femtomole to low picomole |
|
Comments: Excellent LC/MS tool; low salt tolerance (low millimolar); multiple charging useful, but significant suppression with mixtures occurs; low tolerance of mixtures; soft ionization (little fragmentation observed). |
||||||
|
MALDI |
300,000 |
YES |
Photo degradation and matrix reactions |
Good for complex mixtures |
Possible |
Low to high femtomole |
|
Comments: Somewhat tolerant of salts; excellent sensitivity; matrix background can be problem for low mass ions; soft ionization (little fragmentation observed); photodegradation possible; suitable for complex mixtures. Limited multiple charging occurs so MS/MS data is not extensive. |
||||||
References
- Jürgen Cox and Matthias Mann (July 2011). "Quantitative, High-Resolution Proteomics for Data-Driven Systems Biology". Annual Review of Biochemistry. 80: 273–299. doi:10.1146/annurev-biochem-061308-093216. PMID 21548781. – via Annual Reviews (subscription required)
- The mass spectrometer - how it works. https://www.chemguide.co.uk/analysis/masspec/howitworks.html (accessed 04/25/2019)
- McLafferty, F. W. Mass spectrometry across the sciences. Proceedings of the National Academy of Sciences 2008, 105, 18088-18089.
- Bertani, R.; Cecchetto, W.; Polloni, R.; Crociani, B.; Seraglia, R.; Traldi, P. Comparison of hard vaporization and soft ionization techniques in the mass spectrometry of some palladium(II) and platinum(II) complexes with C-bonded heterocyclic ligands. Inorganica Chimica Acta 1990, 174, 61-66.
- Taylor, T. Electron Ionization for GC–MS. LCGC North America 2012, 30,
- Electron ionization.commons.wikimedia.org/wiki/File:Electron_Ionization_-_Born_Oppenheimer_Potential_Curves.png. Wikipedia (accessed 04/25/2019)
- Sparkman, J. T. W. a. O. D.: Introduction to Mass Spectrometry: Instrumentation, Applications, and Strategies for Data Interpretation; John Wiley & Sons, 2007; Vol. 4.
- Barber, M. Fast Atom Bombardment (FAB) & Liquid Secondary Ion Mass Spectrometry (LSIMS). Journal of the Chemical Society 1981, 35.
- . Lafont, H. H. R. Fast-atom bombardment. Reference Module in Life Sciences 2017.
- Barber, M.; Bordoli, R. S.; Sedgwick, R. D.; Tyler, A. N. Fast atom bombardment of solids (F.A.B.): a new ion source for mass spectrometry. Journal of the Chemical Society, Chemical Communications 1981, 325-327.
- Caprioli, R. M.; Farmer, T. B.; Gile, J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Analytical Chemistry 1997, 69, 4751-4760.
- J. B. Fenn, M. M., C. K. Meng, S. F. Wong, and C. M. Whitehouse. Electrospray ionization for mass spectrometry of large biomolecules. Science, 246, 64-71.
- Electrospray ionization. en.Wikipedia.org/wiki/Electrospray_ionization (accessed 04/25/2019).
- Kebarle*, P. A brief overview of the present status of themechanisms involved in electrospray mas spectrometry. JOURNAL OF MASS SPECTROMETRY 2000, 35, 804-817.
- Brackbill, J. U.; Kothe, D. B.; Zemach, C. A continuum method for modeling surface tension. Journal of Computational Physics 1992, 100, 335-354.
- Wilm, M. Principles of electrospray ionization. Molecular & cellular proteomics : MCP 2011, 10, M111.009407-M009111.009407.
- KNOCHENMUSS, Z. A. ION FORMATION IN MALDI MASS SPECTROMETRY. Mass Spectrometry Reviews, 1998, 337–366.
- MALDI-TOF Mass Spectrometry. www.creative-proteomics.com/technology/maldi-tof-mass-spectrometry.htm (accessed 04/25/2019)
- Berkenkamp, S. K., M.; Hillenkamp, F. Role of photoionization and photochemistry in ionization processes of organic moleculesand relevance for matrix-assisted laser desorption/ionization mass spectrometry. Org Mass Spectrom 1992, 30, 1303.
- Calder, A. G., Anderson, S. E., Grant, I. , McNurlan, M. A. and Garlick, P. J. (1992), The determination of low d5‐phenylalanine enrichment (0.002–0.09 atom percent excess), after conversion to phenylethylamine, in relation to protein turnover studies by gass chromatography/electron ionization mass spectrometry. Rapid Commun. Mass Spectrom., 6: 421-424. doi:10.1002/rcm.1290060704
- Julian P, Whitelegge, Stephen M, GómezKym FFaull. Proteomics of Membrane Proteins. Advances in protein chemistry 2003.
- Siuzdak, G. An Introduction to Mass Spectrometry Ionization: An Excerpt from The Expanding Role of Mass Spectrometry in Biotechnology, 2nd ed.; MCC Press: San Diego, 2005. The Scripps Research Institute, Center for Mass Spectrometry 2004.
- M. Barber, R.S. Bordoli, G.J. Elliott, N.J Horoch, and B.N. Green. Biochem. Biophys. Res. Coomun. 110:753-757.
- M. Barber, R.S. Bordoli, R.D. Sedgwick, A. N. Tyler, G.V. Garner, D.B. Gordon, L.W. Tetler, and R.C. Hider. Biomed. Mass Spectrom. 9: 265-269 (1982)
- A. Dell and G.R. Morris. Biochem. Biophys. Res. Commun. 106: 1456-1462 (1982).
- M. Barber, R.S. Bordoli, G.J. Elliott, N.J Horoch, and B.N. Green. J. Chem. Soc. Chem. Commun. 936-938 (1982).
- H. R. Morris, M. Panico, M. Barber, R.S. Bordoli, R.D. Sedgwick, and A.N. Tyler. Biochem. Biophys. RES. Coomun. 101:623-631 (1981).
- Whitelegge, J. P., Gundersen, C. B. and Faull, K. F. (1998), Electrospray‐ionization mass spectrometry of intact intrinsic membrane proteins. Protein Science, 7: 1423-1430. doi:10.1002/pro.5560070619

