2.1: Membrane Fluctuations
- Page ID
- 1355
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)In 1972, Singer and Nicolson [1], proposed a fluid mosaic model for the cell membrane, which viewed the entire membrane as a dynamic system constantly in flux. Since then, the dynamics of cell membranes were studied extensively and have shown that the membrane is in even more flux than the fluid mosaic model had claimed. The membrane is inhomogeneous, with some parts changing their position and composition more rapidly than others. Proteins get inserted and reinternalized, rafts form and dissolve, cytoskeleton is remodeled, and the membrane itself changes its thickness and curvature. Together, all of these perturbations are called membrane fluctuations.
Membrane Composition
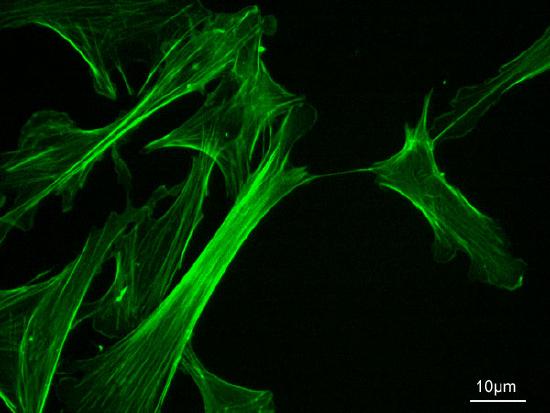
In order to understand the fluctuations that happen within a membrane, a brief overview of the membrane components is required. The membrane consists of a phospholipid bilayer. This bilayer can be thought of as a sea, in which proteins are floating. Proteins can be inserted in either both or a single layer of the membrane. Also in the membrane, there are sterol molecules, which add rigidity. On the inside, the membrane is supported by a cytoskeletal network, which consists mainly of actin (Figure \(\PageIndex{1}\)).

Lipid Rafts
The membrane is not homogeneous; it is full of rafts, or areas of lipids that are in liquid ordered state, which is more ordered than the surrounding area. These rafts are important for a number of reasons; they are thicker than the lesser ordered membrane, and there are transmembrane proteins that only fit in these thicker rafts [2]. These rafts may act as a means to concentrate proteins to an area of the membrane.

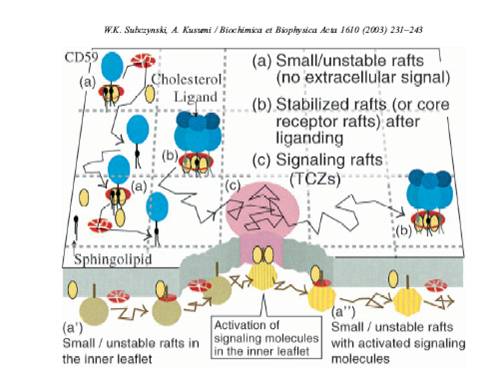
Studies of diffusion rates of raft versus non-raft proteins suggest that the rafts diffuse at a rate similar to that of non-raft transmembrane proteins (Fig 3). This indicates that the rafts are very small, and of similar size to transmembrane proteins.
Proteins
Transmembrane Proteins
There are a multitude of proteins embedded in the membrane. These proteins are continuously recycled, with new proteins being delivered by vesicles. Typically, a transmembrane protein would be synthesized in the rough endoplasmic reticulum (rER) and then transported in a clathrin-coated vesicle to the cell membrane. The vesicle fuses with the membrane, and the proteins become a part of it. Vesicle fusion is a major process of membrane remodeling.
Membrane Remodeling Proteins
There is a number of proteins that can remodel the membrane. One of them is known as BAR (Figure 4) This protein has an inherent curvature, which allows it to sense membrane curvature and induce further curvature by binding. When multiple subunits bind to the membrane, they can cause tubulation.

Actin Network
The cell membrane is anchored to the cytoskeleton, which mainly consists of actin [3], (Fig.5, 6). Actin in itself is a dynamic polymer that is both depolymerizing and polymerizing, depending on the concentration of monomers. This activity is tightly controlled and is extremely important for mobility, phagocytosis, and most processes that require large scale membrane remodeling. By growing actin in a certain point in the cell, membrane can be made to grow protrusions, which can be used to engulf foreign particles or move.

Layer to Layer Fluctuations
It is extremely energetically unfavorable for a lipid from one leaflet of the membrane to flip over to the other. However, this is a process necessary for membrane remodeling and it does occur. A group of enzymes, know as flippases, assists this flipping of lipids [4]. A number of flippases have been identified [5], but the mechanism remains unclear.
Protrusions and Bending
Lipids can move laterally within the membrane, or they can protrude from it. If they form a protrusion, it can be detected with neutron scattering (Figure 7) Bending of the membrane has been suggested to be important in clathrin aggregation using computational methods [6].
. 
References
- Singer SJ, Nicolson GL (Feb 1972). "The fluid mosaic model of the structure of cell membranes". Science 175 (4023): 720– 31
- Jérémie Rossy, Yuanqing Ma, Katharina Gaus, "The organisation of the cell membrane: do proteins rule lipids?", Current Opinion in Chemical Biology, Volume 20, June 2014, Pages 54-59, ISSN 1367-5931
- Mithilesh MishraThe yeast actin cytoskeleton"
- Bretscher, MS. (1972). "Asymmetrical lipid bilayer structure for biological membranes." Nature New Biology 236:11-12
- Islam, S. T. and Lam, J. S. (2013), "Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environmental Microbiology", 15: 1001–1015
- Cordella, Nicholas et al. “Membrane Fluctuations Destabilize Clathrin Protein Lattice Order.” Biophysical Journal 106.7 (2014): 1476–1488

