2.2: Membrane Asymmetry
- Page ID
- 1982
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The purpose of biological membranes is to create compartments suitable for specific biological functions. Biological membranes include a lipid bilayer which functions as a selective permeable barrier to generate internal environments, as well as proteins which participate in molecular transport, catalysis, signaling, cell-cell recognition and forming junctions, and adherence to extracellular spaces. Biomembranes also carry out critical functions in signal transduction and vesicle (de)formation [1].
An asymmetrical membrane lacks uniformity in molecular composition and distribution, and structurally has a variable degree of leaflet curvature. The degree of asymmetry covers a wide range as asymmetry is any deviation of the 50:50 proportion when comparing two surfaces [2]. Asymmetry can occur on both sides of a biological membrane when the inner and outer leaflet have different molecular architecture. One leaflet shows asymmetry (transverse asymmetry) when the molecular components are not homogeneously distributed. Phase separation is one of the simplest examples of transverse membrane asymmetry (Figure \(\PageIndex{1}\)). Lipid rafts are a specific example of membrane asymmetry. They are lateral regions of the bilayer with specific lipid composition that are involved in lipid trafficking and contain proteins participating in various biological functions. The wide variety of lipids, sterols, and membrane-associated proteins evolved correspond to the diverse forms of membrane asymmetry. The loss of physiological membrane asymmetry in cells is deleterious and often results in cell death [18].
This wiki briefly explains the characteristics of the molecular components of membrane bilayers. The contribution of lipids, sterols, and proteins to membrane asymmetry are reflected in specific examples. The last section acknowledges some of the techniques used to assess the presence and degree of membrane asymmetry.
Lipids
Phospholipids
From a historical perspective, lipid asymmetry was first established in the erythrocyte plasma membrane, which has been extensively used as a model for biological membranes [3]. The phospholipid composition in the outer leaflet of the erythrocyte membrane is rich in sphingomyelin and phophatidylcholine (PC). Conversely, the inner leaflet is enriched in phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphatidylserine (PS), all of which form more fluid bilayers than the phospholipids of the outer leaflet [8] (Figure \(\PageIndex{2}\)). Furthermore, PS and PI of the inner leaflet carry a negative charge that interacts with positively-charged residues near the cytoplasmic end of the transmembrane domain of simgle-pass membrane proteins. In some cases, the same phospholipid species can have different acyl chains compared to the opposite leaflet [4, 18]. Remarkably, translocation of phospholipis to maintain the asymmetry in the lipid bilayer is an active process carried out by flippases and floppases via ATP hydrolysis [18].

Nowadays, it is known that the membrane lipids are responsible for the overall cell shape. The charge and size of the head group as well as the number, length and saturation of the acyl chains are all important factors determining membrane symmetry. For example, large, charged head groups are more likely to form curved structures. On the other hand, uncharged lipids with longer acyl chains and smaller head groups would be less likely to form curved structures (Figure \(\PageIndex{3}\)). Thus, local enrichment of specific lipids can induce different curvatures and domains [5]. Lipid molecules tend to group thermodynamically with those that have similar tail length and saturation thus leading to phase separation and domains within the membrane.
Phophatidylserine (PS) on the outer leaflet illustrates the biological importance of maintaining membrane asymmetry. This phospholipid is commonly found in the inner leaflet, but flippases are known to catalyze PS translocation from the inner to the outer leaflet [6]. PS on the extracellular face is known to trigger signaling pathways through activation of ADAM proteins, which cleave transmembrane proteins involved in cell growth and homeostasis. Extracellular PS functions as a tag on apoptotic cells to be phagocyted and participate in platelet activation. A defect in PS translocation to the outer leaflet can result in diseases such as Scott syndrome, where an enzyme, the scramblase, fails to traslocate PS to the outer leaflet as a part of plateler activation [7]. Conversely, Neo1, a P4-ATPase, is a floppase that was recently found to maintain membrane asymmetry translocating PS and phosphatidylethanolamine (PE) to the inner leaflet in yeast [8]. Moreover, translocation of PS to the outer leaflet occurs in erythrocyte adhesion, myotube formation as well as apoptosis, recognition of cells to be phagocyted by macrophages, and malignization of tumor cells [18].
The origin of the lipid asymmetry lays in the organelles of the cell secretory pathway [9]. The endoplasmic reticulum and the Golgi apparatus synthesize and insert the phospholipids specifically in one of the leaflets. Phosphoglycerides, such as phosphatidylcholine, are translocated to the opposite leaflets by flippases and floppases via ATP hydrolysis .
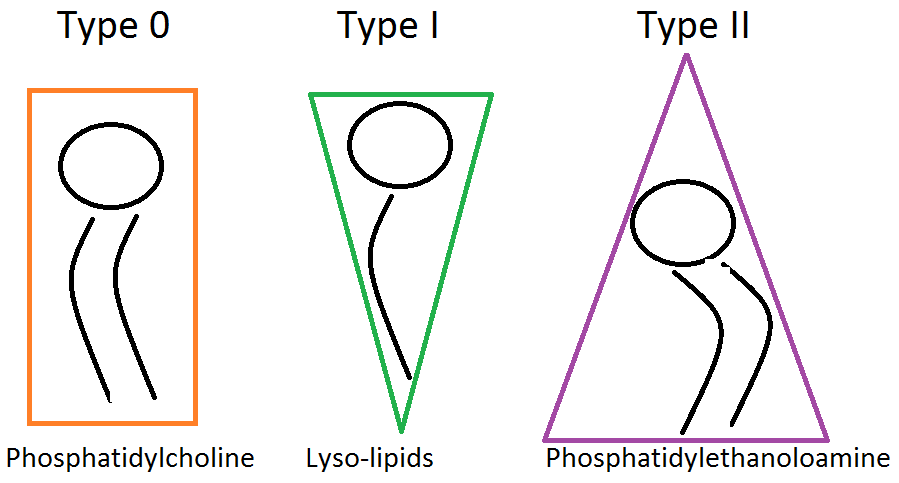
As stated earlier, phospholipids are not the sole reason for membrane asymmetry. Other lipids, carbohydrates and proteins also have a key role in changing the phase and shape of the membrane.
Sterols: Cholesterol
Sterols are four-ring isoprenoid-based hydrocarbons that modulate the fluidity of the lipid bilayers in eukaryotic cells [9]. Among sterols, cholesterol is found in the plasma membrane of animal cells and it is one of the most recently evolved sterols (Figure \(\PageIndex{4}\)). Cholesterol is amphypathic given that is mainly hydrophobic, but also has a hydrophilic hydroxil group.

Cholesterol can be thought of as the moderator of order within the lipid bilayer causing membrane asymmetry. Through its simultaneously bulky head and long tail, cholesterol is able to disorder the gel phase and order the liquid phase of lipids [10]. The presence of cholesterol prevents the phospholipids from being too close to one another, maintaining the membrane fluidity. Likewise, cholesterol provides structural support to the bilayer. The presence of cholesterol buffers the Tm (Transition Temperature) (Figure \(\PageIndex{5}\)) [9]. In lipid rafts, which are in gel phase, the polar head group in cholesterol binds to the amide group in sphingomyelin. This bond prevents the formation of H -bonds between water and cholesterol, which would increase disorder. Cholesterol is also able to order the liquid phase in long-chained lipid-saturated phospholipids in membrane domains. Moreover, cholesterol has recently reported to be a factor that inhibits translocation of PS to the outer leaflet in human erythrocytes, presumably by inhibition of PLSCR1 scramblase. Preventing PS translocation to the extracellular face is important to maintain membrane asymmetry regarding phospholipid composition [11].

The functional role of the membrane proteins and carbohydrates constitute another factor that influences membrane asymmetry. Proteins and carbohydrates tie into the earlier concepts discussed because they interact preferentially with different lipid head groups and tails.
Carbohydrates: Lectins
Carbohydrates are bound to the outer surface of the plasma membrane [3]. This asymmetrical distribution of carbohydrates is related to their functions on the cell surface, such as intercellular adhesion and recognition, and cellular anchorage to the extracellular matrix. Lectins form a group of sugar-binding proteins that participate in cell-cell recognition. Lectins bind to carbohydrates attached to membrane-bound proteins. Glucose, mannose, galactose, N-acetyl galactosamine, sialic acid and fucose are among the most recurrent lectin ligands.
Lipid Rafts
Biological membranes have also lateral asymmetry, evidentially shown by lipid rafts -regions of the outer leaflet involved in specific functions such as lipid trafficking and signaling. The idea of lipid “rafts”, or distinct domains within the membrane, has been proposed as a way to spatially and temporally regulate membrane function [12]. Theoretically, the formation of rafts is thermodynamically favorable. In order to maximize the exclusion of water and increase entropy, acyl chains of similar lengths interact with one another. Similarly, transmembrane (TM) proteins, depending on the length of their TM domain, also preferentially interact with acyl chains of a similar length. The matching of hydrophobic chains and TM domains decreases enthalpy which is even more favorable. The thermodynamic separation of lipids results in phase separations throughout the membrane. The diameter of a lipid raft is 50nm in diameter on average (range: 10-100nm) [4,9].
Lipid rafts are enriched in glycosphingolipids and cholesterol, mainly having saturated acyl chains. As a consequence, lipid rafts are in a liquid-ordered state (Lo), and are thicker than the rest of the bilayer. The uniqueness of the lipid rafts' molecular composition is consistent with their variety of functions [4]. As a matter of fact, polarized cells are known to have specialized domains, like epithelial cells which have apical and basolateral regions. Another example comes from viruses, which only recognize and attach to specific non-random regions in the membrane of the host cells [4]. In addition, only particular membrane domains can participate in cell-cell fusion. Lastly, lipid rafts can be considered as signaling platforms as their proteins are commonly involved in reception and transduction of extracellular signals [9].
Proteins and Membrane Domains
Proteins are asymmetrically distributed across biological membranes according to the different functions carried out in the cytoplasmic and extracellular faces of the membrane [3]. Transmembrane proteins have a specific topology, which is set at the time of their synthesis. As a consequence, the cytoplasmic and extracellular domains can have different structure and function. Most of the extracellular domains of the transmembrane proteins are glycosylated (glycoproteins), and carbohydrates chains are linked to serine, threonine, and asparagine, whereas lipoproteins have lipids attached [9]. Peripheral membrane proteins are anchored to the plasma membrane by electrostatic interactions with either lipids, membrane proteins, or both. In animal cells, a subset of peripheral membrane proteins are linked to the membrane by Glucosyl-phosphatidyl-inositol (GPI) acyl chain on the outer leaflet, and have an important role in signal transduction. In the inner leaflet, membrane proteins are mainly anchored to lipids such as myristic acid, palmitic acid, and prenylated hydrophobic acid chains [3,4].
BAR proteins
Although many proteins are asymmetrically distributed to participate in signaling pathways and catalitic processes, some others have a structural role in maintaining cell shape. Perhaps the best example of protein’s influence on membrane curvature are the Bin-amphiphysin-Rvs (BAR) domain proteins [13]. BAR domains are seen in many important biological context including vesicle and exosome budding. It is no surprise that proteins containing a BAR domain are among the most conserved in biology. BAR domains are banana-shaped dimers that both sense and induce membrane curvature (Figure \(\PageIndex{6}\)). They interact with the negatively charged lipids electrostatically with positively charged residues on one side of their surface, leading to the amphipathic nature of these helical domains.
The computational study in the Figure above was important because it suggested the additive model was correct. This simulation provided some of the first evidence that BAR domains bind linearly along a membrane and that their effects are cooperative [13].
Amphipathic helices
Membrane spanning proteins have an extremely important role in the spatial arrangement of membranes. Local curvature of membranes can be heavily influenced by helical protein insertion [14]. This is true for both anchored and membrane-spanning proteins. The former are typically amphipathic helices, which means they have both a polar and a non-polar face. Non-polar faces typically embed in the membrane and interact with the hydrophobic acyl chains while the polar surface remains near the phospholipid head groups. This kind of asymmetric insertion induces positive curvature in the membrane (Figure \(\PageIndex{7}\)).

One way to study the effects of proteins on membrane curvature is to utilize light microscopy to visualize liposomes or giant lamellar vesicles (GUVs) with and without the presence of protein. With these methods it has been determined that membrane-spanning helices can also manipulate membranes. One common mechanism is by protein oligomerization within the membrane. An example of this membrane manipulation is seen within a specific subdomain of the mitochondrial inner membrane called the cristae junction (CJ). CJs connect the inner boundary membrane to the cristae membranes where oxidative phosphorylation takes place. CJs are highly curved membranes that are responsible for the large surface area of the inner membrane. In fact, Mic10, a subunit of the mitochondrial contact site and cristae organizing system, coordinates the curvature of CJs by oligomerization through its transmembrane helices [15].

Through a glycine-rich oligomerization domain in the transmembrane helices, Mic10 is able to self-assemble and curve membranes (Figure \(\PageIndex{8}\)). Deletion of Mic10 has severe consequences in respiration due to loss of membrane surface area.
Manipulating membrane asymmetry
The membranes in live cells are shown to be inherently asymmetric. Membrane models used for in vitro studies have been difficult to produce asymmetrical bilayers as in living cells [16]. Supported lipid bilayer (SLB) methodology was one of the first approaches aiming to manipulate membrane asymmetry. SLB consist on a lipid bilayer extended on a physical support (surface), which can be a solid, a polymer "cushion" or a self-assembled monolayer [17]. Additionally, SLB can be tethered to the surface, freely- suspended, or a solid-supported lipid bilayer can be the support for vesicular layers (Figure \(\PageIndex{9}\)). The vesicles are initially placed on the surface, and they rupture after support-induced deformation. Ultimately the bilayers from the broken vesicles coalesce and form the SLB (Figure \(\PageIndex{1}\)0). Studies with SLB allow to analyze molecular composition, molecular distribution and the curvature of membranes, which are the factors responsible for membrane asymmetry.

Issues such as bilayer-support unwanted interactions, and the fact that in some cases the supported bilayer becomes completely immobile, account for the difficulties to control membrane asymmetry [16]. Plus, a substantial part of research with SLBs have used vesicles with symmetrical compositions in both leaflets [18]. There are new ongoing approaches in recent years aiming to specifically manipulate membrane asymmetry. These methodologies include droplet-interface bilayers, inverted emulsion techniques, as well as methyl-beta-cyclodextrin catalyzed exchange. The latter approach basically consists of transporting phospholipids from the outer leaflet of a donor liposomes into the outer leaflet of an acceptor liposome [19]. This method is capable to produce asymmetric liposomes in a wide range of diameters, and is considered an easy method of transferring anionic phospholipids such as PS to an outer leaflet of a lipid bilayer, thus resembling membrane asymmetry of biological membranes. In recent years, it has been possible to conduct quantitative studies in lipid flip/flop rates, giving new possibilities for analysis of molecular composition and distribution of biological membranes [18].
References
- Brown, D. A., & London, E. (1998). Functions of lipid rafts in biological membranes. Annual review of cell and developmental biology, 14(1), 111-136.
- Fujimoto, T., & Parmryd, I. (2016). Interleaflet Coupling, Pinning, and Leaflet Asymmetry—Major Players in Plasma Membrane Nanodomain Formation. Frontiers in cell and developmental biology, 4.
- Stillwell, W. (2013). An introduction to biological membranes: from bilayers to rafts. 1e. Academic Press. Elsevier. 367 p.
- Luckey, M. (2014). Membrane structural biology: with biochemical and biophysical foundations. 2e. Cambridge University Press. 411p.
- Harder, T., Scheiffele, P., Verkade, P., & Simons, K. (1998). Lipid domain structure of the plasma membrane revealed by patching of membrane components. The Journal of cell biology, 141(4), 929-942.
- Sommer, A., Bhakdi, S., & Reiss, K. (2016). How membrane asymmetry regulates ADAM17 sheddase function. Cell Cycle, 15(22), 2995.
- Zwaal, R. F., Comfurius, P., & Bevers, E. M. (2004). Scott syndrome, a bleeding disorder caused by defective scrambling of membrane phospholipids. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 1636(2), 119-128.
- Takar, M., Wu, Y., & Graham, T. R. (2016). The essential Neo1 protein from budding yeast plays a role in establishing aminophospholipid asymmetry of the plasma membrane. Journal of Biological Chemistry, 291(30), 15727-15739.
- Lodish, H., Berk, A., Kaiser, C., Krieger, M., Bretscher, A., Ploegh, H., Amon, A., Scott, M. (2013). Molecular cell biology . 7e. New York: Scientific American Books. 1154p.
- Bacia, K., Schwille, P., & Kurzchalia, T. (2005). Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proceedings of the National Academy of Sciences of the United States of America, 102(9), 3272-3277.
- Arashiki, N., Saito, M., Koshino, I., Kamata, K., Hale, J., Mohandas, N., ... & Takakuwa, Y. (2016). An unrecognized function of cholesterol: regulating the mechanism controlling membrane phospholipid asymmetry. Biochemistry, 55(25), 3504-3513.
- Risselada, H. J., & Marrink, S. J. (2008). The molecular face of lipid rafts in model membranes. Proceedings of the National Academy of Sciences, 105(45), 17367-17372.
- Arkhipov, A., Yin, Y., & Schulten, K. (2008). Four-scale description of membrane sculpting by BAR domains. Biophysical journal, 95(6), 2806-2821.
- Zimmerberg, J., & Kozlov, M. M. (2006). How proteins produce cellular membrane curvature. Nature reviews Molecular cell biology, 7(1), 9-19.
- Barbot, M., Jans, D. C., Schulz, C., Denkert, N., Kroppen, B., Hoppert, M., ... & Meinecke, M. (2015). Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell metabolism, 21(5), 756-763.
- Purdie, J. A., & Sanderson, J. M. (2016). Tuning Membrane Asymmetry. Biophysical Journal, 110(3), 85a.
- Richter, R. P., Bérat, R., & Brisson, A. R. (2006). Formation of solid-supported lipid bilayers: an integrated view. Langmuir, 22(8), 3497-3505.
- Marquardt, D., Geier, B., & Pabst, G. (2015). Asymmetric lipid membranes: towards more realistic model systems. Membranes, 5(2), 180-196.
- Markones, M., Zorzin, C., Kalie, L., Fiedler, S., & Heerklotz, H. (2017). Tuning Membrane Asymmetry: Controlled Uptake of Negatively Charged Lipids into the Outer Leaflet of Liposomes. Biophysical Journal, 112(3), 43a.
Supplementary references:
Faller, R., Lecture: Membrane and lipids fundamentals. In: Membrane Biology, UC Davis. April 11th 2017.

