4.3: Protein-lipid Interactions
- Page ID
- 1349
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Lipids and proteins are essential elements of a living organism. Proteins, as one of the most abundant macromolecules of life, have countless functions ranging from catalyzing vital biological reactions to transporting nutrients. Likewise, lipids also play integral roles such as storing energy or being a major component of a membrane [12]. Despite their individual importance, interactions between these two molecules can provide functions that would not be possible individually. The greatest number of these interactions are seen in cell membranes, which are composed of a wide variety of lipids and proteins. While biological membranes elegantly fulfill many structural roles, the complexity of lipid-protein interactions facilitates a potentially stunning diversity of function and interaction.
Background
The crowded biological membrane (Figure \(\PageIndex{1}\)) provides countless interfaces for these interactions. In the membrane, the protein to lipid ratio is believed to be as high as 60:40 [15], and membrane proteins are believed to constitute an approximate 30% of the human genome [5]. These imply not only the individual importance of proteins and lipids to the function of biological membranes, but the potential complexity underlying interactions of proteins and lipids with each other. An understanding of the way proteins and lipids interact thus requires consideration of a variety of factors, from the bulk properties of the membrane to the intermolecular forces contributing to the behavior of individual lipids within the membrane.

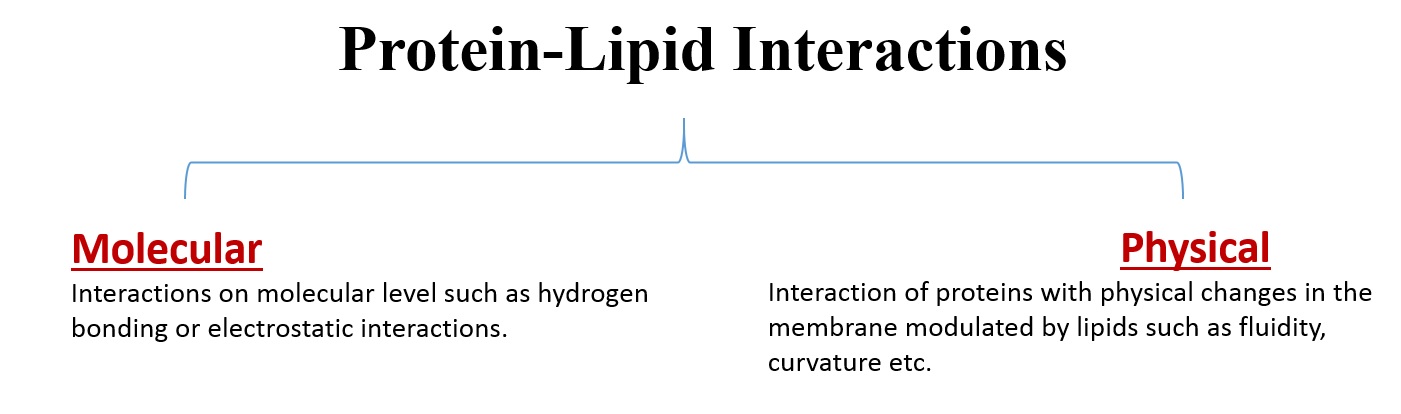
Proteins found in a biological membrane have important roles in cell signaling, transduction and transportation [9]. One common way of classifying protein-lipid interactions is shown below:
Lipids in a biological membrane can be divided into three general groups [9].
- Bulk lipids: The lipids that form the main structure of the membrane without contacting the membrane proteins are called bulk lipids.
- Annular lipids: Some lipids surround membrane proteins and interact relatively non-specifically with them. These lipids are called the annular lipids, due to being similar to an annular shell (a ring-shaped layer in close contact with the protein surface).
- Non-annular lipids: These lipids interact specifically with some membrane proteins and are buried inside the protein structure. Non-annular lipids are often regarded similarly to protein cofactors--small molecules bound by a protein in its functional conformation.
Some of the vocabulary used to discuss membrane and protein properties is summarized in Figure \(\PageIndex{2}\):
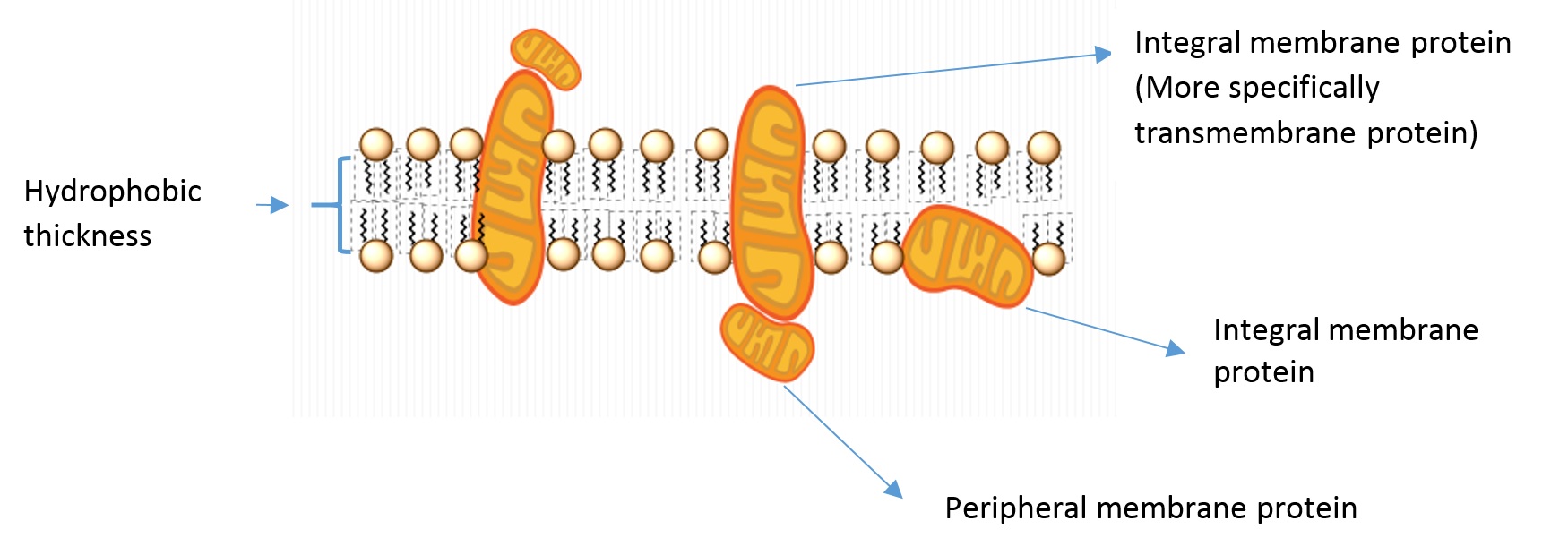
- Integral membrane proteins: Proteins residing in at least one leaflet (layer) of the cell membrane.
- Transmembrane proteins: A class of integral membrane proteins spanning the full width of the cell membrane.
- Peripheral membrane proteins: Proteins that are transiently associated with the outer portion of one leaflet or an integral membrane protein.
- Hydrophobic thickness: The thickness of the portion of the membrane composed of the fatty acid lipid tails.
Bulk Lipid-Protein Interactions
Effect of Viscosity of the Membrane
The movement of any molecule or part of a molecule inside the membrane is influenced by the fluidity of this environment [9]. Every molecule experiences a frictional drag force during their movement inside the lipid bilayer structure of a biological membrane, and resistance of the fluid to this motion can be expressed in terms of the viscosity. By Stokes’ Law (Equation \ref{stokes}), the rate of particle motion can be expressed as a function of the drag force, membrane viscosity, and particle size [3]. This equation assumes the absence of turbulent flow (a good assumption for the comparatively calm cell membrane).
\[v = \dfrac{F}{6π \mu R} \label{stokes}\]
where
- \(F\): drag force (e.g. Newtons)
- \(\mu\): membrane viscosity (e.g. centipoise, Pascal*seconds)
- \(R\): particle size (e.g. meters, nanometers)
- \(v\): speed of particle motion (e.g. meters per second)
Larger particles, more viscous membranes, and more rapid motion thus all result in increased drag force. More viscous membranes can arise from the phase (e.g. liquid disordered, liquid ordered, or gel) of the lipids in the membrane; the phase is determined by the number of double bonds in the fatty acid tails of the lipids and the environmental temperature, with fewer double bonds or a temperature below the critical temperature for the lipids resulting in a more viscous membrane.
Movement of a protein in the membrane is dictated by the frictional resistance and the molecular restoring forces acting on it. A fluorescence polarization anisotropy study on the rotation of tryptophan (Trp) residues showed that the motion of Trp was affected by the viscosity of the lipid environment for small scale motion, and the amplitude of this motion increased with the temperature as the viscosity decreased. For example, at a viscosity of 1 centipoise (approximately the viscosity of water at room temperature and on the order of magnitude of the viscosity of the membrane) the authors expect local rotational motion of a given peptide within the protein to be largely determined by the surrounding peptide residues [17].
While the viscosity of the membrane can affect the rate of motion of the protein, the most thermodynamically favorable conformation of the protein should not depend on membrane viscosity [9]. This is because the thermodynamic properties of a system are inherently time-independent, but viscosity is a time-related parameter (since viscosity has units of pressure*time). This means a change in viscosity cannot directly increase or decrease the activation energy required to induce a conformational change--the most stable conformation of a protein is independent of the membrane viscosity. Changes in membrane viscosity can, however, change the amount of time required for the protein to assume this conformation. Additionally, if a change in membrane viscosity is the result of a change in temperature, the rate of protein function will be affected because rate constants are temperature-dependent [9]. This indirect effect can be worthwhile to consider in the study of isolated membrane proteins, which may take place at room temperature rather than at the temperature of the protein's natural environment.
Another important effect of membrane viscosity has also been revealed by molecular dynamics simulations. It has been illustrated that the effect of solvent viscosity on protein motion is important if the protein both directly contacts the solvent and has a rate of motion comparable to the environment. In other words, high frequency motion of a protein that does not overlap the solvent motion does not depend on the solvent’s viscosity [2]. Lipids are important in this context because viscosity of a biological membrane is mainly influenced by the types of lipids present, as the lipid composition is one of the main determinants of the fluidity.
Highly viscous membranes are indicative of increased ordering of the lipid fatty acid tails, often facilitated by interactions with smaller molecules such as sphingomyelin or cholesterol. This can result in increased membrane durability and impermeability. Additionally, the fluidity of the membrane environment surrounding a membrane protein can impact its function. For example, the human epidermal growth factor receptor (EGFR) was found to be non-functional in an environment in which both highly fluid liquid disordered and more viscous liquid ordered domains could exist. However, in a purely liquid disordered environment, EGFR retained its function, revealing a functional dependence on the fluidity of the membrane [3].
(See the main phase transitions page for more information on membrane fluidity.)
Localized areas of high membrane viscosity can also be indicative of the existence of lipid rafts, which are believed to play a role in protein isolation and function. Lipid rafts are transient sphingomyelin- and cholesterol-rich : (10-200 nm in diameter) within the cell membrane. The local abundance of sphingomyelin and/or cholesterol results in liquid-liquid phase separation, meaning lipid rafts are typically composed of the liquid ordered (Lo) phase. This means the lipids within rafts are more tightly packed and highly ordered than the bulk membrane, leading to increased local rigidity and decreased fluidity [18]. Some proteins can bind glycosphingolipids or sphingomyelin, which are involved in the recruitment of proteins to lipid rafts.
Effect of Membrane Curvature
Dynamic curvature of plasma or intercellular membrane can be dictated by the interactions between proteins and lipids. In fact, cells can employ various mechanisms to sense curvature as a way to create regions of active membrane trafficking. Lipid composition is a major influence on membrane curvature based on their chemical properties and/or the size of their headgroup. Presence of certain lipids are key to interact with certain peripheral membrane proteins in order to induce a necessary curvature. For example, phosphoinositides are required for budding of clathrin-coated vesicles, as the required machinery (e.g. coat proteins) can specifically bind to these lipids. The reason for this is that the headgroups of phosphoinositides are negatively charged, resulting in electrostatic repulsion that can contribute to membrane curvature [14].
Another study illustrated that NSA4(1-48), a key transmembrane protein that mediates replication of Dengue virus (a mosquito born single positive-stranded RNA virus), has an affinity for the convex face of highly curved regions of synthetic bilayer vesicles, as monitored by circular dichroism spectroscopy [8].
Similarly, a G protein-coupled serotonin receptor (5-HT1A) was found to function more rapidly in a more highly curved membrane [7]. This curvature effect could arise from a better match in the thickness of the hydrophobic portion of the membrane surrounding the protein, stabilizing the active protein form.
In addition to responding to changes in local curvature, membrane proteins can also influence the local curvature of the membrane. Some membrane proteins accomplish this by trafficking individual lipids across the membrane, resulting in uneven lipid compositions across the two membrane leaflets. The membrane must curve so that the leaflet with fewer lipids is on the interior of the curve in order to maintain optimal packing of the head groups [6]. Similarly, membrane leaflets containing lipids with different head group areas may also have some intrinsic curvature due to their differing surface areas [6]. In both cases, the integrity of the membrane as a barrier is facilitated by curvature, protecting the hydrophobic fatty acid tails from the aqueous environment and maintaining separation between the membrane bound compartment and its exterior.
Yet another example of the interplay between membrane proteins and membrane curvature is the process of vesicle formation. The ability to form vesicles is important in the trafficking of newly-formed proteins, the recycling of cell membrane components, and the intake of particles via endocytosis, among countless other cell functions. Vesicle formation involves complex regions of curvature, including low positive curvature when the vesicle is first beginning to form, greater positive curvature as the vesicle swells, and very high negative curvature where the vesicle meets the parent membrane. This complex curvature is facilitated in part by the membrane protein dynamin, which is responsible for creating the “neck” separating the parent membrane from the nearly fully-formed vesicle. This is a result of dynamin binding the membrane and taking on a helical conformation, physically forcing the parent membrane to assume a tube-like shape [14]. Coatomers are small proteins that may also play a role in vesicle formation by binding existing membrane proteins in a relatively confined area. The coatomers result in steric repulsion, leading to membrane curvature and budding [6].
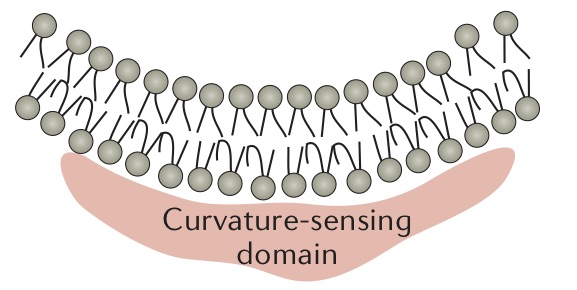
The binding of amphipathic helices within the membrane is another common mechanism of protein-induced membrane curvature. As shown in the Figure below, one portion of the helix is hydrophobic, while the other is hydrophilic. The hydrophobic portion associates with the fatty acyl chains of the membrane, while the hydrophilic portion interacts with the head groups. This changes the area per head group for the bound leaflet of the membrane, and the membrane must curve in order to compensate.
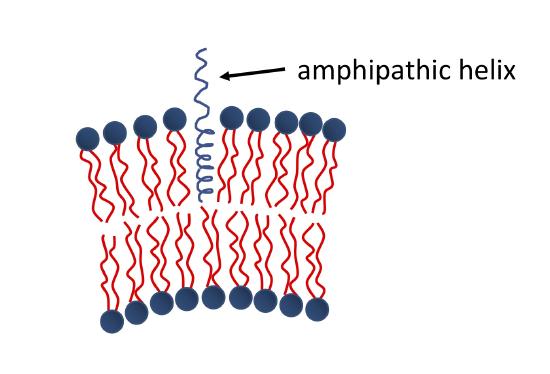
Some of the other well understood ways that lipid-protein interaction relates to curvature of the membrane are discussed in membrane curvature in detail.
Effect of Changes in Membrane Thickness
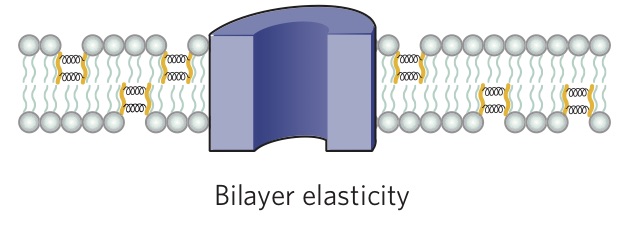
An important feature of lipids that can influence protein function is the thickness of the bilayer. As shown in Figure \(\PageIndex{2}\), the hydrophobic thickness of a membrane is the distance between the hydrophilic headgroups of either leaflet, which is related to the length of the fatty acid tails of the lipids composing the membrane. Membrane proteins need to match the hydrophobic thickness of the acyl chains for two reasons. First, acyl chains and hydrophobic groups of the membrane proteins do not form hydrogen bonds with water, and tend to minimize their contact with it as this is more thermodynamically favorable (see hydrophobic effect). When the hydrophobic thickness of the membrane and the membrane protein do not match, hydrophobic mismatch occurs. In order to minimize the exposure of hydrophobic portions of the lipid bilayer and transmembrane proteins to the aqueous environment, the bilayer can distort in various ways. It can stretch (Figure \(\PageIndex{5}\)), compress, or even tilt. Stretching involves an extension of the fatty acid chains of the lipids immediately surrounding the protein, providing a greater hydrophobic thickness to better match the size of the protein's hydrophobic domain. This minimizes the extent of hydrophobic membrane protein exposed to the aqueous environment, which is more energetically favorable. Conversely, compression involves a contraction of the fatty acid tails surrounding the protein.
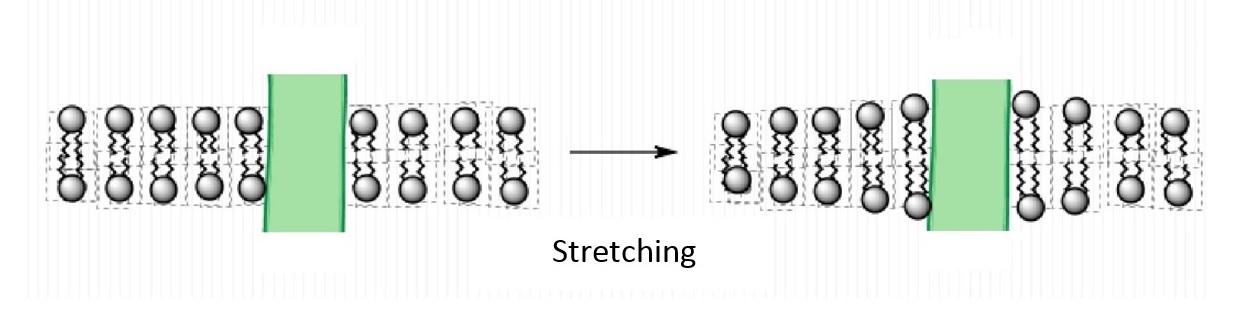
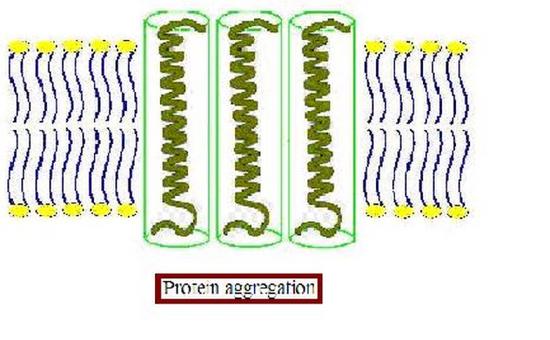
Furthermore, the membrane proteins can aggregate (Figure \(\PageIndex{3}\)) to minimize the area exposed to water [16]. In addition to being more thermodynamically favorable (i.e. maximizing entropy by reducing the number of water molecules in highly ordered contact around hydrophobic residues), avoiding exposure of a transmembrane protein to aqueous environment is also important for optimal function because the hydrophobic side chains of the protein can change conformation when they contact water, distorting their native structure, and causing functional loss. The studies on these membrane distortions carried out with model membranes (artificial bilayers), show that the thickness of the acyl chain region is an essential part of protein function. For instance, sarcoplasmic reticulum Ca-ATPase, which is a protein important in calcium regulation in muscles, was shown to be greatly affected by the number of the carbons in the acyl chain, which determines the hydrophobic thickness of the membrane [16]. In addition, molecular dynamics simulations showed the possibility of protein aggregation (Figure \(\PageIndex{3}\)) due to hydrophobic mismatch [17].
Membrane proteins may also change conformation in response to changes in membrane tension, which can be indicative of changing external conditions necessitating a response from the cell. For example, osmotic swelling or shrinking of a cell will result in higher or lower membrane tension, respectively. Under increased tension, the bilayer will tend to thin, resulting in increased hydrophobic mismatch. One possibility to mitigate this is for the membrane conformation to change so that its hydrophobic domain thickness matches that of the transmembrane protein. Another possibility is that a conformational change in the membrane protein structure could relieve hydrophobic mismatch, making a response to membrane tension an effective gating mechanism. This is typically more energetically favorable in the case of significant membrane deformations, for which the energy required to change the membrane conformation to match the protein’s hydrophobic thickness would be greater than the cost of changing the protein’s conformation [11]. Such conformational changes include changes in the angle at which the transmembrane domain is tilted or unwinding of alpha-helical transmembrane domains [11]. The response of membrane proteins to hydrophobic mismatch arising from membrane tension is evident in the case of many bacterial mechanosensitive channels, which under higher membrane tension are more likely to be open [15].
For example, the lipid-protein interaction energy for a membrane protein with six helical transmembrane domains has been estimated to increase by approximately 10-20 J/lipid molecule for a 10 Å hydrophobic mismatch, providing an appreciable driving force for either the membrane or protein to change its conformation [11]. At room temperature, the energetic cost of bending a lipid membrane is typically on the order of 10-20 J, although this can change significantly based on the local temperature and lipid composition [24]. Hydrolyzing one ATP, a typical means of driving protein conformational changes, yields approximately 10-19 J [6]. All these values are within similar orders of magnitude, suggesting the most effective method of resolving hydrophobic mismatch for a given membrane and protein may depend on both intrinsic and environmental factors.
Annular and Non-Annular Lipid-Protein Interactions
Protein-Lipid Interactions in Molecular Terms
In addition to the interactions of lipids and proteins mediated by the physical changes in the lipid environment, it is good to consider lipid-protein interactions on a molecular level. Types of lipids that were defined previously interact differently with proteins. Figure \(\PageIndex{4}\) is a basic representation of the interaction of bulk lipids and annular lipids with a protein surface. Lipids present in the first shell, having a higher level of interaction with the protein and more restricted motion, are the "annular lipids.” The second shell shows the bulk lipids.

Annular lipids and proteins weakly interact through van der Waals interactions, hydrogen bonding, and/or electrostatic interactions. These weak interactions allow annular lipids to be frequently exchanged with the lipids in the bulk bilayer due to the random motion expected in any fluid system, which implies that their interactions with proteins are fairly nonspecific. The level of interactions between annular lipids, which are highly dynamic, is an ongoing area of research as it still is a challenging task to analyze their structure. One example of interactions of such lipids with membrane proteins suggests that charges of annular lipids are important to the functionality of certain proteins. Accordingly, a type of ABC transporter protein, which can be involved in diseases such as multidrug resistance, preferred interacting with phosphatidylglycerol, a negatively charged phospholipid, and not with phosphatidylethanolamine, a zwitterionic phospholipid. This suggested the presence of higher affinity spots on the protein [1].
The neuronal protein syntaxin-1A, involved in the release of neurotransmitters, has also been shown to interact preferentially with negatively charged lipids [23]. Syntaxin-1A is found primarily in segregated clusters due to the local excess of a particular anionic lipid. The electrostatic interactions between the protein and lipid facilitate the formation of a protein microdomain without a change in the local phase of the surrounding lipids [23].
The existence of an annular shell of lipids around the transmembrane domain of a membrane protein arises from the distortion of the lipid fatty acid tails at the lipid-protein interface. The introduction of a solid, geometrically complex surface—the protein—into the membrane forces a rearrangement of the surrounding fatty acid tails in order to maintain the impermeability of the membrane. Without the lipid annulus, the tails would be able to pack only poorly around the protein, resulting in membrane leakiness. The entropic penalty of maintaining the ordered annular lipid shell is thus worthwhile in order to preserve the structural integrity of the membrane [15].
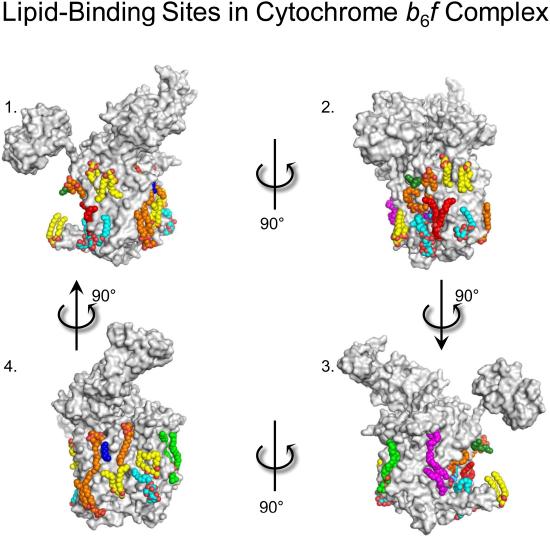
Unlike bulk and annular lipids, non-annular lipids interact specifically with proteins. These lipids are buried inside the protein, and are thought to be vital for optimal function. An example of such an interaction is found to be useful in bovine heart cytochrome c oxidase, on which 13 different types of lipids were suggested to have specific binding sites. It was also suggested that two palmitate tails of phosphatidylglycerols have a role in blocking the O2 transfer pathway of this protein [19]. Lipid binding sites on the exterior of a similar protein are shown in the Figure below.
The interaction of non-annular lipids with proteins may also have implications in the detergent solubilization of membrane proteins—a common method used to isolate proteins from their native membranes to better understand the protein’s structure or function. If the detergent used is sufficiently similar to the annular lipids typically surrounding the protein, there is a possibility that the detergent could penetrate this lipid shell and interact with the protein during the solubilization process [13]. This could change the conformation of the protein, potentially impacting its function. For example, the amino acid transporter LeuT has been shown to bind detergent molecules in one of its active sites (which displace the lipids typically residing there). This reduces the activity of LeuT. However, when a larger detergent molecule was used, it was unable to penetrate the annular lipid shell during the solubilization process, resulting in a retention of the protein’s function [13]. This indicates that an understanding of the annular lipids in a protein's native membrane can be crucial to preserving both its structure and its function if solubilized.
Experimental Methods Used to Study Lipid-Protein Interactions
Some of the techniques used to study physical properties of lipid environment interacting with the membrane proteins include fluorescence spectroscopy, and electron paramagnetic resonance (EPR). There are methods utilized to understand interactions in molecular terms based on resolving the three-dimensional structures of proteins, which also helps clarify functions of some of the non-annular lipids [20]. X-Ray crystallography, NMR or EPR are among common tools analyzing lipid-protein interactions in molecular terms. Molecular dynamics simulations are another area of focus that can help researchers understand interactions of proteins and lipids both in molecular and physical terms.

The isolation of membrane proteins in proteoliposomes has also become a relatively common method of studying membrane protein function, including lipid-protein interactions. A proteoliposome is a small spherical vesicle into which a protein has been inserted. Proteoliposomes can be formed with known lipid compositions, allowing study of the effect of lipid composition--impacting membrane viscosity, phase separation, and membrane thickness-- on a protein’s function. The membrane of the proteoliposome can thus act as a host to a variety of membrane proteins. While this method cannot necessarily replicate the inherent crowding and complexity of a native cell membrane, it can provide an understanding of the relative membrane properties required for a protein’s function or the ligand affinity of the protein, among other things.
References
- Bechara, C., Noll, A., Morgner, N., Degiacomi, M., Tampe, R., & Robinson, C. (2015). A Subset of Annular Lipids is linked to the Flippase Activity of an ABC Transporter. Biophysical Journal.
- Brooks, C.L.; Karplus, M. Solvent effects on protein motion and protein effects on solvent motion. J. Mol. Biol. 208 (1989) 159–181.
- Coskun, Ü.; Grzybek, M.; Drechsel, D.; Simons, K. Regulation of Human EGF Receptor by Lipids. Proc. Natl. Acad. Sci. 2011, 108 (22), 9044–9048.
- Deen, W. Analysis of Transport Phenomena, 2nd ed.; Oxford University Press, 2011.
- Fagerberg, L.; Jonasson, K.; von Heijne, G.; Uhlén, M.; Berglund, L. Prediction of the Human Membrane Proteome. Proteomics 2010, 10, 1141–1149.
- Faller, R., MCB/BPH 241 Course Lectures. 2019.
- Gutierrez, M. G.; Mansfield, K. S.; Malmstadt, N. The Functional Activity of the Human Serotonin 5-HT1A Receptor Is Controlled by Lipid Bilayer Composition. Biophys. J. 2016, 110 (11), 2486–2495.
- Hung, Y.;Schwarten, M.; Schünke, S.; Thiagarajan-Rosenkranz, P.; Hoffmann, S.; Sklan, E.; Koenig, B. (2015). Dengue virus NS4A cytoplasmic domain binding to liposomes is sensitive to membrane curvature. Biochimica Et Biophysica Acta (BBA) - Biomembranes, 1119-1126.
- Lee, A.G. (2004). How lipids affect the activities of integral membrane proteins. Biochimica Et Biophysica Acta (BBA) - Biomembranes, 62-87.
- Lee, A.G. (2011). Biological membranes: The importance of molecular detail. Trends in Biochemical Sciences, 493-500.
- Lee, A. G. Lipid-Protein Interactions. Biochem. Soc. Trans. 2011, 39 (3), 761–766.
- Lehninger, A. (1982). Principles of biochemistry. New York, N.Y.: Worth.
- LeVine, M. V; Khelashvili, G.; Shi, L.; Quick, M.; Javitch, J. A.; Weinstein, H. Role of Annular Lipids in the Functional Properties of Leucine Transporter LeuT Proteomicelles. Biochemistry 2016, 55 (6), 850–859
- McMahon, H. T.; Gallop, J. L. Membrane Curvature and Mechanisms of Dynamic Cell Membrane Remodelling. Nature 2005, 438 (7068), 590–596.
- Phillips, R.; Ursell, T.; Wiggins, P.; Sens, P. Emerging Roles for Lipids in Shaping Membrane-Protein Function. Nature 2009, 459, 379–385
- Razvan, L., & Thomas, D. (1994). Effects of Membrane Thickness on the Molecular Dynamics and Enzymatic Activity of Reconstituted Ca-ATPase. Biochemistry, 33, 2912-2920.
- Rholam, M., Scarlata, S., & Weber, G. (1984). Frictional resistance to the local rotations of fluorophores in proteins. Biochemistry, 6793-6796.
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The Mystery of Membrane Organization: Composition, Regulation and Roles of Lipid Rafts. Nat. Rev. Mol. Cell Biol. 2017, 18 (6), 361–374
- Shinzawa-Itoh, K. et al. (2007) Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713– 1725
- Sperotto, M.M. A theoretical model for the association of amphiphilic transmembrane peptides in lipid bilayers. Eur. Biophys. J., 26 (1997), pp. 405–416
- Hydrophobic mismatch. (2013, September 20). In Wikipedia, The Free Encyclopedia. Retrieved 21:05, May 13, 2015, from en.Wikipedia.org/w/index.php?title=Hydrophobic_mismatch&oldid=5737297
- Engelman, D. M. Membranes Are More Mosaic than Fluid. Nature 2005, 438 (7068), 578–580.
- van den Bogaart, G.; Meyenberg, K.; Risselada, H. J.; Amin, H.; Willig, K. I.; Hubrich, B. E.; Dier, M.; Hell, S. W.; Grubmüller, H.; Diederichsen, U.; et al. Membrane Protein Sequestering by Ionic Protein–Lipid Interactions. Nature 2011, 479 (7374), 552–555.
- Palivan, C. G.; Goers, R.; Najer, A.; Zhang, X.; Car, A.; Meier, W. Bioinspired Polymer Vesicles and Membranes for Biological and Medical Applications. Chem. Soc. Rev 2016, 45, 377–411.
- Harayama, T.; Riezman, H. Understanding the Diversity of Membrane Lipid Composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296.
- Hasan, S. S.; Cramer, W. A. Internal Lipid Architecture of the Hetero-Oligomeric Cytochrome B6f Complex. Structure 2014, 22 (7), 1008–1015.

